Research
Epigenetics / Chromatin Chemical Biology
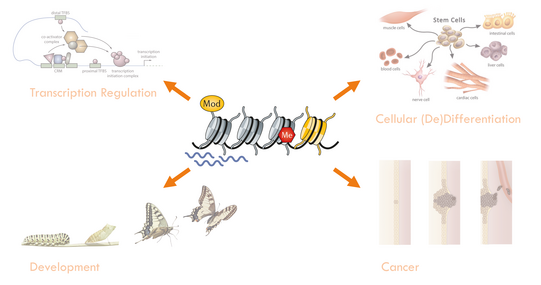
The regulated expression of genes underlies virtually any biological process ranging from cell differentiation and development to the onset of diseases such as cancer. The majority of this regulation takes place at the level of chromatin.
However, the mechanistic picture of this regulation is increasingly complicated by the continuous discovery of numerous regulatory elements that act in concert to dynamically control the structure/function and thus transcriptional activity of chromatin. These include reversible decorations of DNA, RNA and nuclear proteins with regulatory chemical marks, noncoding RNA transcripts, and long-range chromatin interactions.
While the discovery and mapping of such elements become increasingly straightforward, the understanding of their precise mechanistic functions in chromatin regulation remains largely incomplete, since detailed structural and functional studies of the involved protein-nucleic acid complexes in an unperturbed, cellular environment are hampered by a lack of suitable methodology.
Our group is focused on studying such regulatory elements in chromatin by chemical biology approaches. We devise novel strategies to reengineer and/or control basic molecular properties of nucleic acid-interacting proteins to enable novel insights into chromatin regulation. We thereby combine chemical and biological methodologies including organic synthesis and biomolecular chemistry, genetic code expansion, directed molecular evolution, imaging, and high throughput genomic analyses.
A current focus of the lab is the role of epigenetic DNA modifications in the regulation of protein-DNA complex formation in chromatin both at the local and system-wide level. To study these roles, we
1.) Evolve new classes of reader proteins that offer unconventional strategies for interrogating DNA modifications via in vitro and cellular analyses. Such readers also have the potential to be used for epigenome engineering with previously inaccessible selectivity. We further employ directed evolution strategies to study the selectivity of natural chromatin proteins for specific settings of DNA modifications on the proteome level.
2.) A second field of research is the development of novel optochemical tools that allow precise temporal and spatial control of the readers, writers and erasers of DNA modifications in cells, and thus enable dissecting the order and kinetics of downstream events that lead to altered chromatin states and transcriptional activities under normal and disease conditions.