Research
Research Concept
In our research group we investigate how cells perform their functions by controlling their dynamic shape. We particularly focus on the mechanisms that underlie the organization of dynamic, fibrous structures that are collectively called the cytoskeleton. To gain insight into these mechanisms, we first identify key system components. We then interrogate causalities between these components by combining acute activity perturbations and live-cell imaging. On the basis of these experiments, we build mathematical models of the spatio-temporal system dynamics, which help us to generate new, testable hypotheses.
Current Research
Dynamic shape changes in cell migration
During embryonal development, wound healing or during the immune response, cells have to navigate complex tissues to fulfill their function. To enable their movements, cells dynamically alter their shape, either by pushing themselves outward to generate protrusions, or by pulling themselves back via retractions. To effectively reach their destination, cells have to coordinate these protrusions and retractions in space and time. How cells achieve this coordination is still poorly understood. In our recent studies [1] we focused on two signal network components that act as central components in the regulation of cell migration: Rac1, which stimulates cell protrusion and RhoA, which stimulates cell retraction. Contrary to previous models, we found that active Rac1 can stimulate RhoA and that this functional link can locally couple cell protrusion and cell retraction events inside individual cells [1]. Modulation of the underlying mechanism showed that strong coupling between Rac1 and RhoA activity is responsible for dynamic cell shape changes and that this leads to uncontrolled, non-directional migration, which is typically observed in transformed cancer cells [1]. Video 1 shows a representative cell that undergoes such dynamic shape changes, highlighting increased activity of Rac1 just at the leading edge during the phase of cell protrusion.
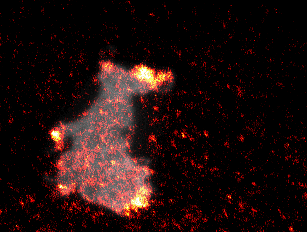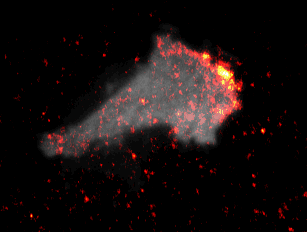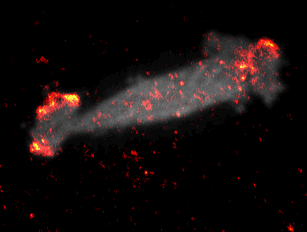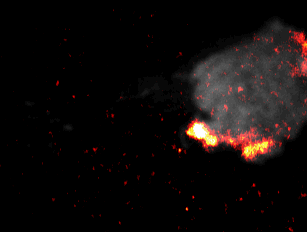
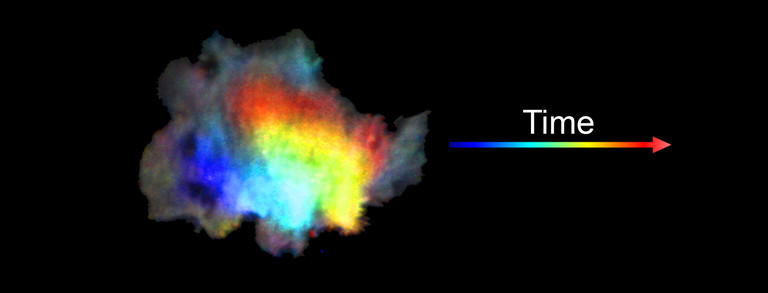
Video 1: Video microscopy of a single cancer cell that dynamically changes its migration direction. The active form of the regulator Rac1 (bright red) is strongly enriched at the cell edge during the protrusion phase and absent during cell retraction.
[1] Nanda S, Calderon A, Sachan A, Duong TT, Koch J, Xin X, Solouk-Stahlberg D, Wu YW, Nalbant P, Dehmelt L. (2023)
Rho GTPase activity crosstalk mediated by Arhgef11 and Arhgef12 coordinates cell protrusion-retraction cycles.
Nat Commun 14:8356
Self-organization of cell contraction: "A sense of touch for individual cells"
In the context of our studies on cell contraction, we uncovered a self-organizing mechanism that leads to the spontaneous emergence of local pulses and propagating waves of cell contraction and of the associated signal network component Rho (Video 2) [1,2]. Our experimental analysis showed that Rho rapidly amplifies its own activity by recruiting its activator GEF-H1 and that it inhibits its activity via slow activation of the contraction-inducing molecular motor myosin II and associated regulators. This combination of a fast positive and slow negative feedback loop is responsible for the generation of the observed local pulses and propagating waves of Rho activity shown in Video 2. Furthermore, activity dynamics are modulated by matrix elasticity, showing that extracellular mechanical cues are coupled with the signal network that generates cell contraction pulses. Thus, cells use such cell contraction pulses to locally squeeze the plasma membrane and to probe the elasticity of their surroundings, and they use this information to modulate their behavior. Individual cells therefore have a sense of touch that uses an active probing mechanism, which is based on the local, subcellular control of signal network activity.

Video 2: Propagation of self-amplified and self-inhibited activity that controls the contraction of the plasma membrane of cells. Here, the signal molecule is the small GTPase Rho, which can exist in an active or inactive state. Specifically, Rho regulates cell contraction by activation of a molecular motor called myosin. The video shows a time-lapse of a single, human cancer cell. Bright and warm colors represent high activity levels of Rho.
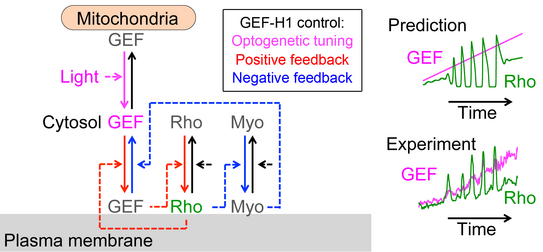
More recently, we combined acute optogenetic perturbations and monitoring of signal network activity responses to derive the reaction-diffusion system that generates pulses and waves of cell contraction [2]. Quantitative experimental and theoretical investigations of this system revealed that the cytosolic concentration and diffusion of the Rho activator GEF-H1 is a critical parameter, that controls Rho activity patterns and how they are modulated by mechanical inputs [2] (Figure 1).
Relevant Literature
[1] Graessl M, Koch J, Calderon A, Kamps D, Banerjee S, Mazel T, Schulze N, Jungkurth JK, Patwardhan R, Solouk D, Hampe N, Hoffmann B, Dehmelt L, Nalbant P (2017).
An excitable Rho GTPase signaling network generates dynamic subcellular contraction patterns.
J Cell Biol 21(14):5311-6.
doi: 10.1083/jcb.201706052.
[2] Kamps D, Koch J, Juma VO, Campillo-Funollet E, Graessl M, Banerjee S, Mazel T, Chen X, Wu YW, Portet S, Madzvamuse A, Nalbant P, Dehmelt L, (2020).
Optogenetic Tuning Reveals Rho Amplification-Dependent Dynamics of a Cell Contraction Signal Network.
Cell Reports 33(9), 108467
Acute perturbation and activity measurements in living cells
To uncover mechanisms, how cellular structures are organized in space and time, methods are required that enable direct monitoring and acute perturbation of key regulators. To reach this goal, we developed novel generic approaches to simultaneously analyze and modulate biochemical reactions inside living cells.
Protein interaction arrays in living cells
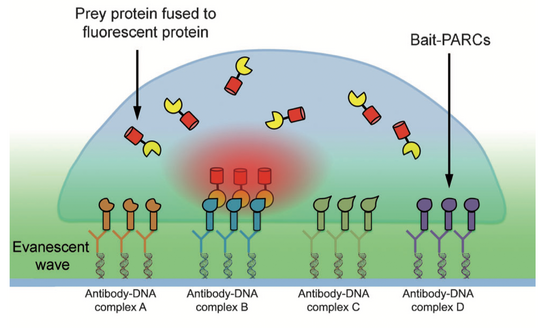
In particular, relations between multiple protein reactions have to be measured simultaneously inside individual cells to untangle complex signal networks. However, current technologies to analyze protein reactions in cells are limited by the small number of markers that can be distinguished via microscopy. To break this barrier, we developed miniaturized protein arrays that allow simultaneous monitoring of multiple protein interactions inside individual living cells (Figure 2) [1]. We are currently applying this technology to study signal networks that control cell shape changes.
Figure 2: Protein arrays inside living cells. Bait presenting artificial receptor constructs (bait-PARCs) transfer an antibody surface pattern into an ordered array of intracellular bait proteins. The interaction of a labeled prey protein with multiple bait proteins is monitored inside living cells via microscopy.
Relevant Literature
[1] Gandor S, Reisewitz S, Venkatachalapathy M, Arrabito G, Reibner M, Schröder H, Ruf K, Niemeyer CM, Bastiaens PI, Dehmelt L (2013).
A protein-interaction array inside a living cell.
Angew Chem Int Ed Engl 52(18):4790-4.
doi: 10.1002/anie.201209127.
"Molecular Activity Painting": Switch-like, light-controlled perturbations inside living cells
To induce acute and prolonged perturbations of protein activities in the plasma membrane we developed methods based on chemically-induced dimerization and photochemically-induced targeting to immobilized artificial receptors to directly “paint” stable network perturbations in living cells (Video 3) [4]. To combine those perturbations with activity measurements, we developed TIRF-based methods to measure the activity of the major Rho GTPases Rac1, Cdc42 and RhoA. Using these tools, we directly investigated perturbation response relationships in the spatio-temporal processing of cell contractility signaling.
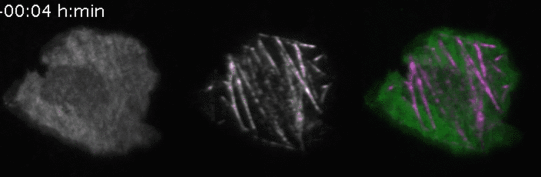
Video 3: Molecular activity painting of the letter “N” via ~1µm wide lines of the Rho activitor GEF-H1 (left panel). Plasma membrane localization of GEF-H1 induced the new formation of dynamic, myosin-based contractile structures (middle panel). Right panel: combined channels.
Relevant Literature
[3] Chen X, Venkatachalapathy M, Kamps D, Weigel S, Kumar R, Orlich M, Garrecht R, Hirtz M, Niemeyer CM, Wu YW, Dehmelt L. (2017). "Molecular-Activity Painting": Switch-like, Light-Controlled Perturbations inside Living Cells. Angew Chem Int Ed Engl 21(14):5311-6.
doi: 10.1002/anie.201611432