Research
Current research undertaken in Kasanmascheff Lab revolves around understanding the in vitro and in vivo structure, function and dynamics of biomolecules and bioorganic radicals that play key roles in life such as biosynthesis of DNA building blocks, higher-order DNA G-quadruplex structures, FeS proteins and proton-coupled electron transfer reactions. To reach our goals, we extensively employ EPR spectroscopy combined with sophisticated biochemical and biophysical techniques.
In vivo structure determination of biomolecules
The function of biomolecules is dictated by their structure and dynamics, which are affected by the protein environment. In vitro experiments might differ from the in vivo ones where molecular crowding, protein-protein interactions and the concentration of co-solutes may play significant roles on the structure and conformational heterogeneity of proteins. Therefore, in our lab we study biomolecules in their natural environment, the biological cell.
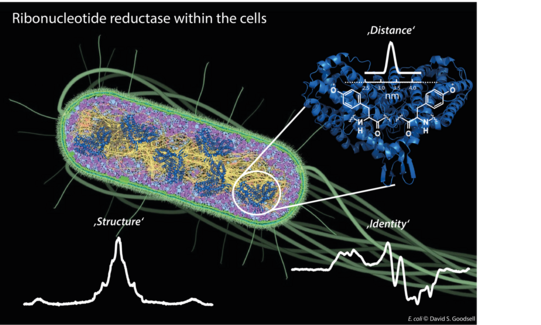
Our first target is the E. coli ribonucleotide reductase (RNR) that catalyzes the biosynthesis of DNA building blocks in E. coli and humans. RNRs are a paradigm to study radical transfer steps in essential protein machineries and also well-recognized targets for cancer therapeutics. Therefore, determining its in vivo structure and regulation is of high significance (see research highlights for more detail).
Figure 1: Different EPR techniques are employed to give insights into in vivo structure and function of E. coli ribonucleotide reductase.
Higher-order DNA G-quadruplex structures and their formation kinetics
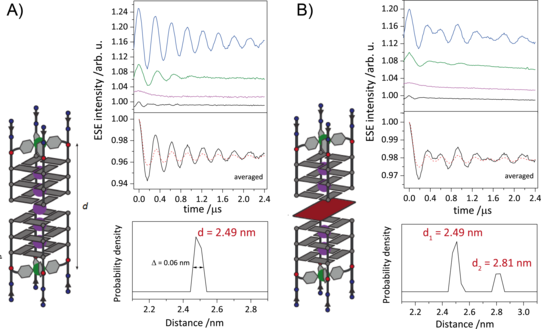
G-quadruplexes are DNA secondary structures containing stacked guanine tetrads stabilized by central cations. Studies have shown that G-quadruplexes form in vivo in oncogene regulatory regions and at telomeric ends of chromosomes, thus shortening cancer cell lifetimes. Many G-quadruplex species are known to form higher-order structures like dimers, which are believed to play a crucial role in G-quadruplexes biological activity. This makes the understanding of the formation of these structures an important goal. We use EPR spectroscopy to gain insight into the topology of these higher-order structures by deriving CuII–CuII distances in various forms of G-quadruplex dimers, in which Cu(pyridine)4 complexes are covalently incorporated into tetramolecular G-quadruplexes. With the CuII-based spin labels attached at either 3’ or 5’ ends of several G-quadruplex species, we show the dimerization at the opposite ends and follow the kinetics of heterogenous dimerization via dipolar spectroscopy.
Figure 2: A) Schematic representation of DNA G-quadruplex dimers (right). Orientation selective distance measurements with background-corrected orientation-averaged distance measurement (upper left) and corresponding distance distribution (lower left). B) Schematic representation of sandwich complex of DNA G-quadruplex and free guanine tetrad (left). Orientation selective distance measurements with background-corrected orientation-averaged distance measurement (upper right) and corresponding distance distributions (lower right). The two distances represent the DNA G-quadruplex dimers and the sandwich complex.
EPR-monitored redox potentiometry
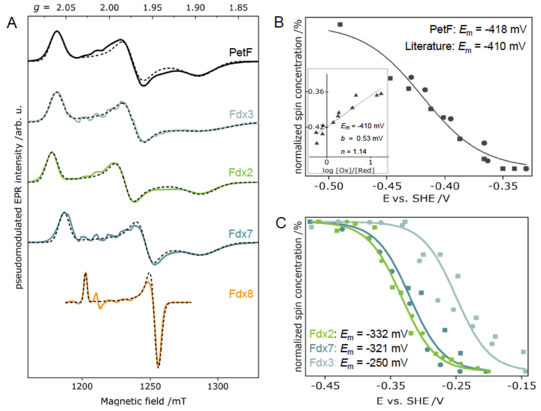
Determination of redox potentials of FeS proteins provide essential information on the energetics of biological electron transfer reactions. Therefore, we built an electrochemical cell for mediated titrations of metalloproteins, and thus obtaining their redox potentials via EPR-monitored potentiometric titrations. We established a protocol for the accurate determination of the redox potentials via pulse EPR for the first time. We started determining the redox potentials of ferredoxins (Fdxs), which are small, soluble FeS proteins. Additionally, we develop strategies to tune the midpoint potentials of selected ferredoxin, i.e., point mutations of charged residues on the protein surface and/or in vicinity of the FeS clusters.
Figure 3: (A) Electron-spin echo EPR spectra of different plant-type Fdx isoforms and their corresponding simulations. Plots of normalized spin concentrations against the respective potentials of the reductive (circles) and oxidative (squares) redox potentiometries of PetF (Fdx1) and Fdx2, Fdx3 and Fdx7.
Light-activated radicals
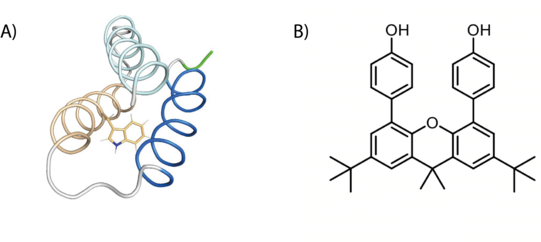
Tyrosine (Y) and tryptophan (W) residues can serve as one-electron redox cofactors in biocatalysis and multistep proton-coupled electron transfer (PCET) reactions. Enzymes that employ amino-acid radical cofactors are involved in essential processes in primary metabolism such as photosynthesis, respiration, and biosynthesis of DNA building blocks. Therefore, detailed understanding of amino-acid radical mediated PCET reactions is of vital interest. In order to investigate the amino-acid radical meditated PCET, we utilize light activation and EPR spectroscopy on two distinct systems: a model protein that is specifically designed to study amino-acid radicals, and a synthetic system that mimics the tyrosine dyad in RNR (DPX).
Figure 4: Schematic representation of an amino-acid radical and DPX (B).